Building Better Genomics Tools Together
Returning home to a community of genomics tool builders at the 2025 Nextflow Summit in Boston
May was a busy month, and I’m catching up with blog posts! Here’s a backlog post from a conference in May for scientists using the niche programming language Nextflow, originally developed for genomics.
At Seanome, we’re building open-source tools that figure out the “jobs” of proteins in the cell, unlocking the forgotten 99% of life on Earth. The vast majority (far greater than 99%) of proteins have unknown functions, and we’re changing that by building Kmerseek to identify more novel protein functions, faster.
Being part of the nf-core community is key to Seanome’s success because we want to engage with the global community of tool and pipeline builders. We are developing our own tool for annotating unknown proteins (Kmerseek) but to do that, we need to (1) benchmark it against the competition to demonstrate Kmerseek can find things other tools can’t and (2) empower OTHERS to use it as easily as possible! Hence, we are building nf-core/proteinannotator, a unified Nextflow pipeline to characterize “mystery” proteins, aka proteins of unknown function from their sequence data.
To engage with the global community of pipeline builders, I attended the 2025 Boston Nextflow Summit, a 4-day event from May 13-16. It was divided into 2 days of training or hackathon, and 2 days of Summit talks.

It was fun reconnecting with people I hadn’t seen since the last Nextflow event I attended — back in 2019 when it was called “Nextflow Camp!”
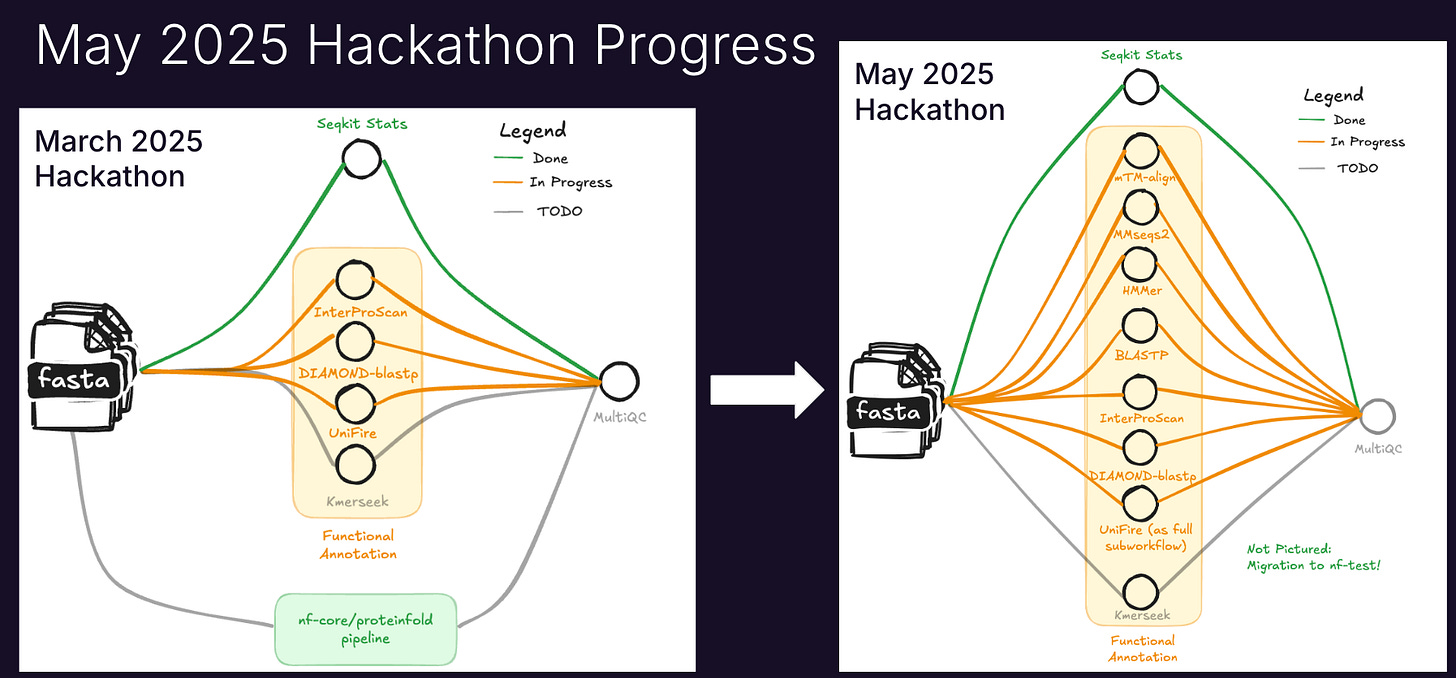
For the first two days, I led a hackathon group for nf-core/proteinannotator. The goal is for someone who sequenced a cool new creature to use our pipeline to figure out what their proteins do!
We made a bunch of progress: hackathon attendees added 4 tools to the pipeline! Key ones were mTM-align, MMseqs2, HMMer, and of course, BLASTp. Thanks to Chase, Addy, and Jessica for helping! Integration of the tools into the pipeline is still in progress and help is welcome!
Seqera was kind enough to give me a speaking slot where I talked about Seanome and how we’re building the best protein annotator in the world! It was fun to do this back-to-back with the hackathon and update the community on our progress. Check out the recording of the talk on YouTube.
I’m excited for more progress on nf-core/proteinannotator! This week I merged a long-standing pull request to add InterProScan to the pipeline, which I started at the March 2025 nf-core Hackathon.
If you want to help out with Seanome, the best place to contribute by picking an issue on the nf-core/proteinannotator GitHub repo and get coding!